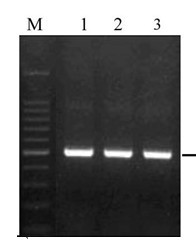
Beijing adlai providing a stool extraction Genomic DNA was extracted from the silica gel membrane technology fresh or frozen human feces or other type of sample (mixed with high concentrations of PCR inhibitors) in the genome of bacteria, viruses, and to send multiple of 30 μ g Biological DNA . A unique adsorption resin with optimized buffer removes PCR inhibitors. Aidlab convenient purification spin columns 40 minutes, up to 30 μ for purification from genomic g feces, bacteria, parasites, and the DNA viruses; Rapid Purification of high quality, ready to use the DNA; no organic extraction or ethanol precipitation; Length High yield; complete removal of contaminants and downstream reaction inhibitors. The purified DNA can be up to 50 kb in length . This length of DNA is fully denatured and has the highest amplification efficiency. High purity DNA can be used directly in downstream amplification reactions. Principle, method, and operation are consistent. The perfect replacement for Qiagen 's QiaAmp DNA Stool Mini Kit . v         product description: Conventional methods of DNA purification are not effective in removing large amounts of inhibitory factors present in feces and leading to failure of downstream experiments, such as PCR, which does not amplify the desired fragment. The kit uses a DNA adsorption column and a new unique solution system to effectively remove various inhibitors in animal feces that affect downstream experiments (such as PCR ) and efficiently recover genomic DNA from feces . After the animal feces sample is resuspended in a special buffer ASL , v         Features: 1.          The silicon matrix membranes in the centrifugal adsorption column are all made of special adsorption membranes imported by world famous companies. The difference in adsorption between columns and columns is very small and the repeatability is good. Overcome the shortcomings of the quality of the domestic kit membrane. 2.          It is not necessary to use reagents such as toxic phenol, and does not require steps such as ethanol precipitation. 3.          Fast, simple, and single sample operation is typically completed in 40 minutes. 4.          Multiple column rinses ensure high purity. The typical ratio of OD 260 / OD 280 is 1.7 ~ 1.9 , which can be directly used for PCR , Southern-blot and various digestion reactions. 1.          All centrifugation steps are done at room temperature using conventional benchtop centrifuges up to 13,000 rpm , such as Eppendorf 2.          Preheat the required water bath before starting the experiment. 3.          Binding fluid CB And the inhibitor-removing fluid IR contains irritating compounds, wear latex gloves during operation to avoid skin, eyes and clothing. If it is contaminated with skin or eyes, rinse with plenty of water or saline. 4.          The eluent EB does not contain the chelating agent EDTA . Does not affect downstream enzyme digestion, ligation and other reactions. It can also be eluted with water. However, it should be ensured that the pH is greater than 7.5 and the pH is too low to affect the elution efficiency. DNA eluted with water should be stored in - 5.          PCR may be inhibited by excess DNA (generally greater than 1 μg ), and a better amount of eluted DNA (appropriate dilution) may be used to obtain better amplification. The volume of eluted DNA that is typically added should not exceed 10 % of the total PCR reaction volume . We recommend adding a final concentration of 0.1 μg/μl of BSA (bovine serum albumin) to the PCR reaction system to help achieve optimal amplification. v         Operation steps: (Please read the precautions before the experiment) 1.          Collect about 200-220 mg of feces into a 2 ml centrifuge tube and place on ice. If it is a frozen specimen, it cannot be thawed before adding the buffer ASL , otherwise the DNA is easily degraded. 2.          Add 1.4 ml of buffer ASL and vortex continuously for 1 minute or until homogenously homogenized. Be careful to mix thoroughly by vortexing, otherwise the yield will be severely reduced. 3.          Resuspension This heating step can increase 3-5 times DNA yield and help lyse bacteria and parasites. For some cells that are difficult to lyse (such as Gram-positive bacteria) can be improved to 4.          Vortex for 15 seconds and let stand for 1 minute at room temperature . The fecal pellets were precipitated at the highest speed for 1 minute. 5.          Transfer 900 μl of the supernatant to a 1.5 ml centrifuge tube, add 100 μl of the impurity scavenger AB , and immediately vortex for 1 minute or until completely homogenized, and let stand for 1 minute at room temperature . The impurities were removed by centrifugation at high speed for 3 minutes. 6.          Transfer all supernatant to a 1.5 ml centrifuge tube and centrifuge at maximum speed for 3 minutes. 7.          Transfer 210 μl of the supernatant to a 1.5 ml centrifuge tube, add 20 μl of proteinase K (20 mg/ml) solution, mix well, add 200 μl The combined solution CB was vortexed for 15 seconds and thoroughly mixed. If the yield is low, more supernatant can be transferred and the corresponding amount of proteinase K and binding solution and subsequent isopropanol use can be increased proportionally . 8.          Add 100 μl after cooling Isopropanol, vortex and mix. 9.          The solution obtained in the previous step and the precipitate which may be present are added to an adsorption column AC (the adsorption column is placed in a collection tube) and centrifuged at 13,000 rpm for 30 seconds, and the waste liquid in the collection tube is discarded. 10.       500 μl of inhibitor-removing solution IR was added , centrifuged at 12,000 rpm for 30 seconds, and the waste liquid was discarded. 11.       Add 700 μl of rinse WB (please check if absolute ethanol has been added ! ), centrifuge at 12,000 rpm for 30 seconds, and discard the waste. 12.       500 μl of the rinse WB was added , centrifuged at 12,000 rpm for 30 seconds, and the waste liquid was discarded. 13.       Place the adsorption column AC back into the empty collection tube and centrifuge at 13,000 rpm for 2 minutes to remove the rinse solution as much as possible. In order to avoid residual ethanol in the rinse solution, the downstream reaction is inhibited. 14.       Remove the adsorption column AC , put it into a clean centrifuge tube, add 100 - 150μl elution buffer EB in the middle of the adsorption membrane (elution buffer can be in advance at 65 The larger the elution volume, the higher the elution efficiency. If the DNA concentration is required to be high, the elution volume can be appropriately reduced. However, the minimum volume should not be less than 50μl , and the volume is too small to reduce DNA elution efficiency and reduce DNA yield. 15.       DNA can be stored in 2 Description: genomic DNA extracted faecal stool Aidlab genomic DNA, followed by PCR amplification using a primer enteropathogens. Lane M : 100 bp DNA Ladder ; Lanes 1-3 : PCR results for detection of intestinal pathogens . Medical protective glasses,Anti-epidemic medical goggles,Isolated medical goggles,Quality medical protective glasses Hebei Abiding Co.Ltd , https://www.hebeiabiding.com v         Precautions
How to quickly extract genomic DNA from feces (no inhibitor, can be directly PCR)
How to quickly extract genomic DNA from feces (no inhibitor, can be directly PCR ) Keywords: fecal DNA extraction   Fecal genome extraction   Genomic DNA extraction